Damon Clark, Ph.D.
Associate Professor of Molecular, Cellular & Developmental Biology, of Physics, and of Neuroscience
Director of QBio Institute
damon.clark@yale.edu
Our lab is interested in how networks of neurons perform computations. The fruit fly’s
visual-motor transformations serve as an ideal model in which we can follow neural
processing as the brain uses visual stimuli to guide behaviors. We want to understand how
animals extract information from the complex spatiotemporal visual patterns in the natural
world, and how they make decisions based on that information. By comparing fly algorithms
and mechanisms to vertebrate ones, we look for evolutionary constraints and diversity in
solutions to basic computational tasks.
Visit the
Clark Lab website.
Thierry Emonet, Ph.D.
Associate Professor of Molecular, Cellular & Developmental Biology, and of Physics
thierry.emonet@yale.edu
Our lab combines mathematical modeling and quantitative experiments to understand the biological computations that enable organisms to sense and navigate their chemical environments. Chemical navigation involves many non-trivial computations and therefore provides a quantitative framework for discovering how biological systems compute, and how computations are implemented in molecular and cellular mechanisms. As model systems, we use the well characterized bacterial chemotaxis and fly olfaction systems. The dual perspective of microbiology and neuroscience helps reveal general principles while fostering innovation by cross-pollinating ideas.
Visit the Emonet Lab website.
Joe Howard, Ph.D.
Eugene Higgins Professor of Molecular Biophysics & Biochemistry, and of Physics
jonathon.howard@yale.edu
We work on motor proteins and the cytoskeleton and how they organize, shape and move cells. We are a highly interdisciplinary group with biologists, physicists, chemists and engineers. By combining optical, mechanical and biochemical experiments with theory and computation we are trying to understand how motor proteins and microtubules function as molecular machines and how they contribute to such cell biological processes as mitosis, ciliary and flagellar motility, mechanosensing and the morphogenesis of neurons.
Visit the Howard Lab website.
Roy Lederman, Ph.D.
Assistant Professor of Statistics and Data Science
roy.lederman@yale.edu
We are interested in the mathematics of data science, inverse problems and unsupervised learning, Bayesian inference and variational inference, numerical analysis, and applied harmonics analysis. Our work is motivated by applications in cryo-electron microscopy (cryo-EM).
Visit Roy Lederman's website.
Christopher Lynn, Ph.D.
Our research aims to understand the statistical mechanics of the brain (and other complex living systems). In neuroscience and biology, microscopic interactions build upon one another to produce macroscopic behaviors and impressive feats of information processing. To understand how these large-scale phenomena emerge from fine-scale interactions, we build upon ideas from statistical physics, information theory, and network science. Using these insights, we investigate how collective patterns of activity and structure arise in neural systems, and study how these patterns encode, communicate, and compress information.
lynnlab.yale.edu
Benjamin Machta, Ph.D.
Our research focuses on understanding how biological systems operate using approaches from theoretical physics. Our group uses statistical physics to understand the structure and function of biological membranes, which experiments have shown operate close to a demixing critical point in the Ising universality class. We also use tools from information theory to place physical bounds on the ability of organisms to function that arise from their limited ability to measure their environment and from their limited energy budget. Lab site: https://machtagroup.yale.edu/.
Visit the Machta Group website.
Alison Sweeney, Ph.D.
Associate Professor of Physics; Ecology and Evolutionary Biology
alison.sweeney@yale.edu
Our lab focuses on biological soft matter and its evolution. We are particularly interested in the evolutionary mechanisms underlying episodes in which the evolutionary process discovers a novel mode of self-assembly. Recent examples of this include their work on self-assembly of gradient-index lenses in squids, and micron-scale surface textures in pollen.
Visit Alison Sweeney's website.
Paul Turner, Ph.D.
Rachel Carson Professor of Ecology and Evolutionary Biology
paul.turner@yale.edu
The main focus of our lab group is to study evolutionary genetics and genomics of microbes, especially the ability of viruses to adapt (or not) to changes in their biotic and abiotic environments. These studies concern environmental challenges faced by viruses at all levels of biological organization, including effects of changes in molecules, proteins, cells, populations, communities and ecosystems. Our work is highly interdisciplinary, employing microbiology, computational biology, genomics, molecular biology and mathematical-modeling approaches, and especially experimental evolution (‘evolution-in-action’) studies under controlled laboratory conditions.
Visit the Turner Lab website.
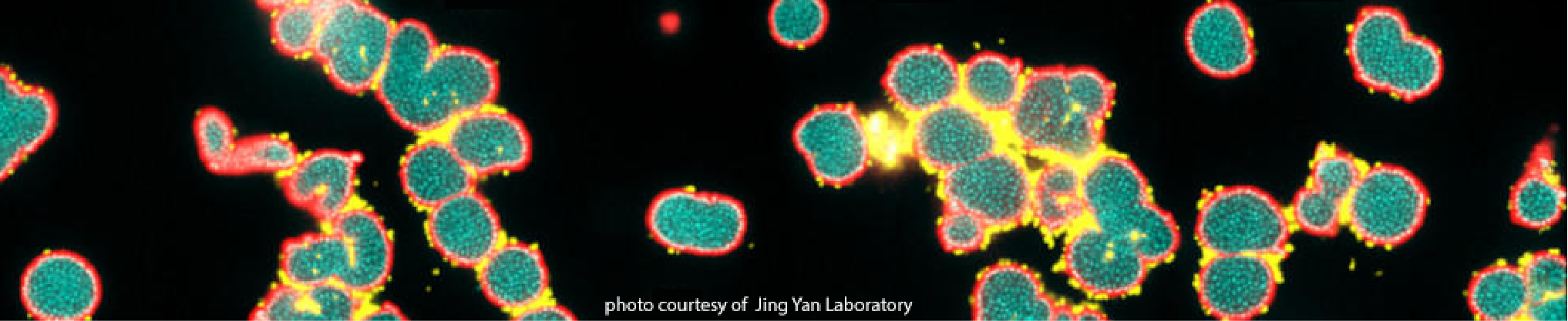
Jing Yan, Ph.D
Assistant Professor, Molecular, Cellular and Developmental Biology
jing.yan@yale.edu
We are a discovery-driven research group working at the interface between microbiology, molecular biology, physics, and engineering. In particular, we are interested in the formation of biofilms, surface-attached bacterial communities embedded in an extracellular matrix. We combine state-of-art imaging techniques, mutagenesis, mechanical measurements, and computer simulations to understand how bacteria build such multicellular communities cell by cell, what unique materials they use to do so, and what characteristics emerge at the level of the collective. Ultimately, we will use our understanding of bacterial biofilms to solve biofilm-related problems in medicine and in industry and to enhance the use of beneficial biofilms.
Visit the Yan Lab website or
Jing Yan's personal website.