The Quantitative Biology Institute at Yale (QBio) serves as the University’s central hub for research and education at the intersection of biology, physics, mathematics, engineering, and computer science. QBio seeks to understand how biological systems solve problems and how the structure, organization, and behavior of living systems emerge from such problem-solving. By decoding the problem-solving algorithms shaped by evolution, QBio aims to reveal the logic of life and train the next generation of interdisciplinary scientists.
Explore the people and the research highlights of QBio faculty members:
News
-
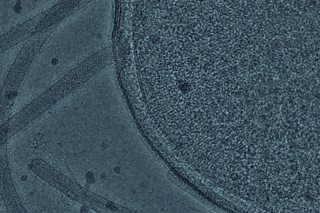
Mapping of brain mitochondria
Recent Science paper from Damon Clark’s group is featured on Insights & Outcomes of YaleNews.
-
E. coli chemosensing accuracy is not limited by stochastic molecule arrivals
Collaboration between Emonet and Machta Labs uncovers new insights into bacterial chemotaxis.
-
New View of Cholera ‘Tail’
YSM researchers and Jing Yan’s Lab used a new microscopy techniques to reveal the molecular structure of the flagella that could inform new treatment.
Funding
The faculty members of the Yale Quantitative Biology Institute gratefully acknowledge the financial support from several funding agencies, including Alfred P. Sloan Foundation, Gordon & Betty Moore Foundation, Human Frontier Science Program, National Science Foundation (NSF), National Institutes of Health (NIH), The Pew Charitable Trust, US Department of Energy, Yale University, etc.